Spatial Interpolation - Part 1¶
Analysis Preparation¶
Imports¶
All of these libraries should have been previously installed during the environment set-up, if they have not been installed already you can use install.packages(c("raster", "rgdal", "ape", "scales", "deldir", "gstat"))
library(raster) # rasters
library(rgdal) # spatial data
library(sf) # for handling spatial features
library(ape) # clustering (Morans I)
library(scales) # transparencies
library(deldir) # triangulation
library(gstat) # geostatistics
source('../scripts/helpers.R') # helper script, note that '../' is used to change into the directory above the directory this notebook is in
Loading Data¶
We’ll start by again checking to see if we need to download any data.
download_data()
We’ll then load in the datasets.
df_zambia <- read_sf('../data/zambia/zambia.shp')
df_africa_cities <- read_sf('../data/africa/cities.shp')
solar <- raster('../data/zambia/solar.tif')
altitude <- raster('../data/zambia/altitude.tif')
Do some pre-processing on the zambian cities dataset (which we’ll use as points in our Voronoi diagram later).
df_zambian_cities <- df_africa_cities[which(df_africa_cities$COUNTRY=='Zambia'), ]
df_zambian_cities_UTM35S <- st_transform(df_zambian_cities, crs=st_crs(20935))
head(df_zambian_cities_UTM35S, 3)
| LATLONGID | LATITUDE | LONGITUDE | URBORRUR | YEAR | ES90POP | ES95POP | ES00POP | INSGRUSED | CONTINENT | geometry | ⋯ | SCHADMNM | ADMNM1 | ADMNM2 | TYPE | SRCTYP | COORDSRCE | DATSRC | LOCNDATSRC | NOTES | geometry |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <chr> | <dbl> | <int> | <int> | <int> | <chr> | <chr> | ⋯ | <chr> | <chr> | <chr> | <chr> | <chr> | <chr> | <chr> | <chr> | <chr> | <POINT [m]> | |
| 4 | -12.65000 | 28.08333 | U | 1990 | 9945 | 10684 | 11218 | N | Africa | POINT (617646.2 8601614) | ⋯ | COPPERBELT | Copperbelt | Kalulushi | UA | Published | NIMA | Census of Population, Housing & Argriculture 1990 Descriptive Tables | UN Statistical Library | NA | POINT (617646.2 8601614) |
| 7 | -12.36667 | 27.83333 | U | 1990 | 48055 | 51629 | 54206 | N | Africa | POINT (590593.6 8633048) | ⋯ | COPPERBELT | Copperbelt | Chililabombwe | UA | Published | NIMA | Census of Population, Housing & Argriculture 1990 Descriptive Tables | UN Statistical Library | NA | POINT (590593.6 8633048) |
| 9 | -12.53333 | 27.85000 | U | 1990 | 142383 | 152972 | 160610 | N | Africa | POINT (592346.6 8614610) | ⋯ | COPPERBELT | Copperbelt | Chingola | UA | Published | NIMA | Census of Population, Housing & Argriculture 1990 Descriptive Tables | UN Statistical Library | NA | POINT (592346.6 8614610) |
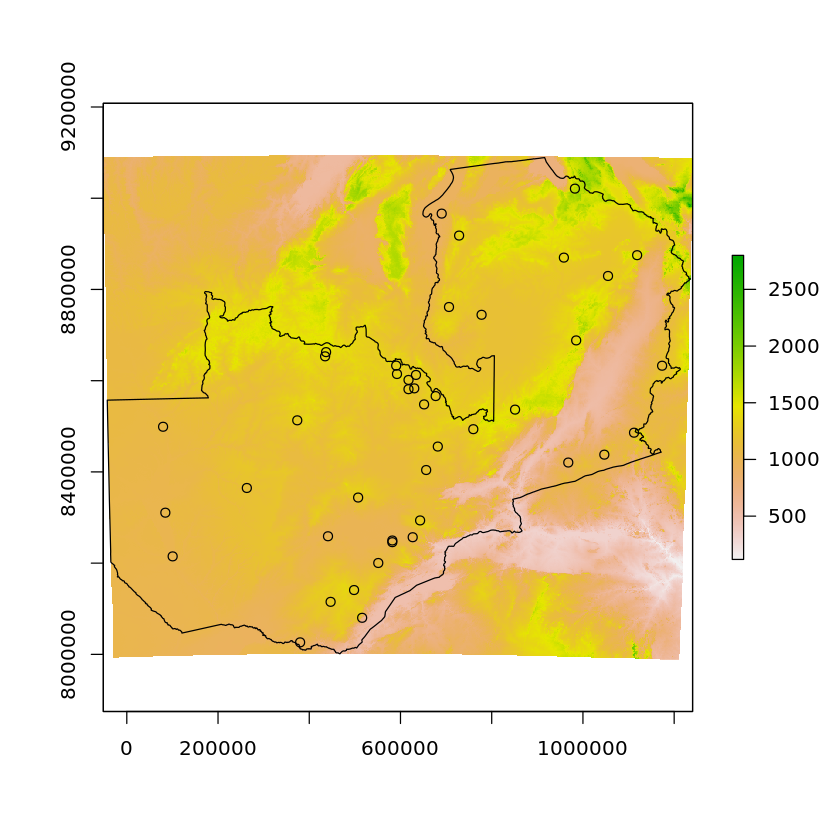
We’ll visualise these as well
plot(altitude)
plot(df_zambia['geometry'], add=TRUE)
plot(df_zambian_cities_UTM35S['geometry'], add=TRUE)
Voronoi Tesselation/Thiessen Polygons¶
Before we move on to interpolation we’ll look at how we can create a Voronoi diagram:
… a partitioning of a plane into regions based on distance to points in a specific subset of the plane. That set of points (called seeds, sites, or generators) is specified beforehand, and for each seed there is a corresponding region consisting of all points closer to that seed than to any other. - Wikipedia
We’ll now use the deldir function to carry out Delaunay triangulation and Dirichlet tessellation, this function returns a complex object.
cities_deldir <- deldir(df_zambian_cities_UTM35S$LONGITUDE, df_zambian_cities_UTM35S$LATITUDE)
cities_deldir
Delaunay triangulation and Dirchlet tessellation
of 43 data points.
Enclosing rectangular window:
[22.11,34.190001] x [-18.751667,-7.931666]
Area of rectangular window (total area of
Dirichlet tiles):
130.7056
Area of convex hull of points (total area of
Delaunay triangles):
54.96517
We can use tile.list to convert the complex object returned in the last step into a list where each entry is a separate Dirichlet/Voronoi tile
cities_polygons <- tile.list(cities_deldir) # Produces Thiessen polygons
plot(cities_polygons)
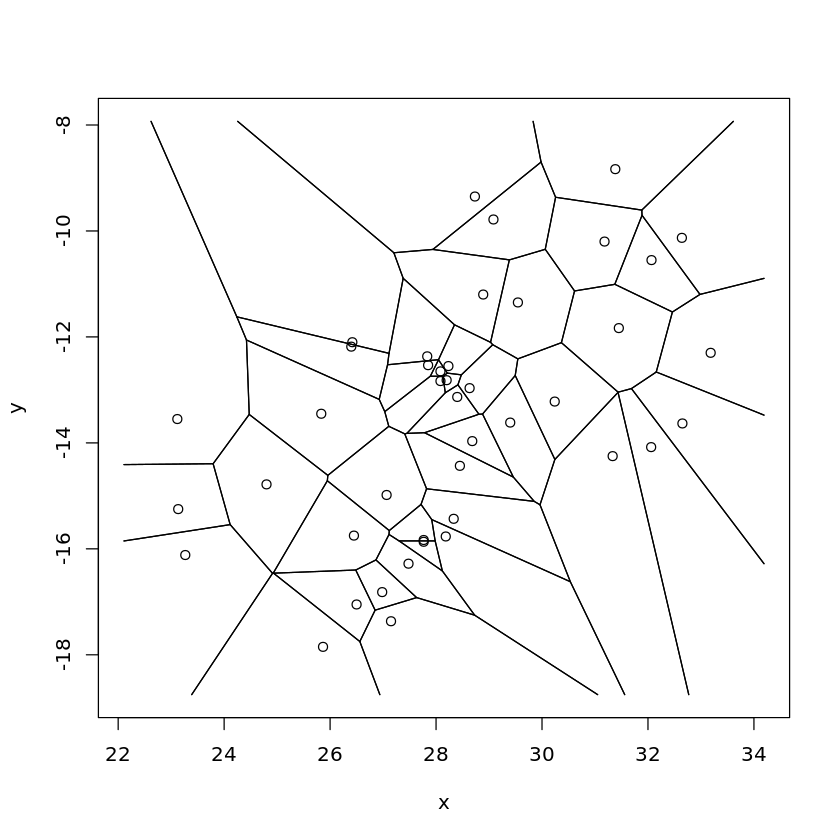
We can then susbset the returned object, with each element relating to a single city in the original dataset, the area attribute is the size of the Polygon for that city.
cities_polygons[[1]]
- $ptNum
- 1
- $pt
- x
- 28.083334
- y
- -12.65
- $x
-
- 28.046429
- 27.891667
- 28.129763
- 28.213637
- $y
-
- -12.432142
- -12.741666
- -12.741666
- -12.682955
- $bp
-
- FALSE
- FALSE
- FALSE
- FALSE
- $area
- 0.052275
Interpolation¶
Before we start interpolating we’ll quickly inspect the dataset for any clear trends, starting with the lon/lat & altitude relationship with solar potential.
First we’ll extract the solar and altitude values for the points we’ve selected (just so happens to be cities in this case).
city_coords = st_coordinates(df_zambian_cities_UTM35S)
df_zambian_cities_UTM35S$solar <- extract(solar, city_coords)
df_zambian_cities_UTM35S$altitude <- extract(altitude, city_coords)
df_zambian_cities_UTM35S$x <- city_coords[, 1]
df_zambian_cities_UTM35S$y <- city_coords[, 2]
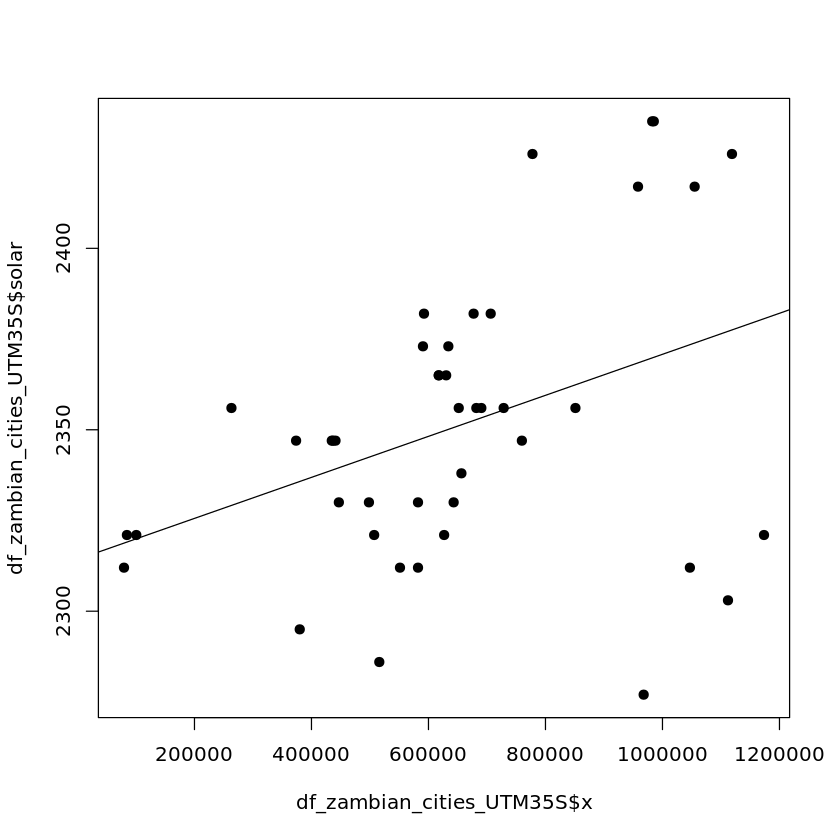
Longitude appears to have a weak relationship with solar output
plot(df_zambian_cities_UTM35S$x, df_zambian_cities_UTM35S$solar, pch=19)
abline(lm(df_zambian_cities_UTM35S$solar ~ df_zambian_cities_UTM35S$x))
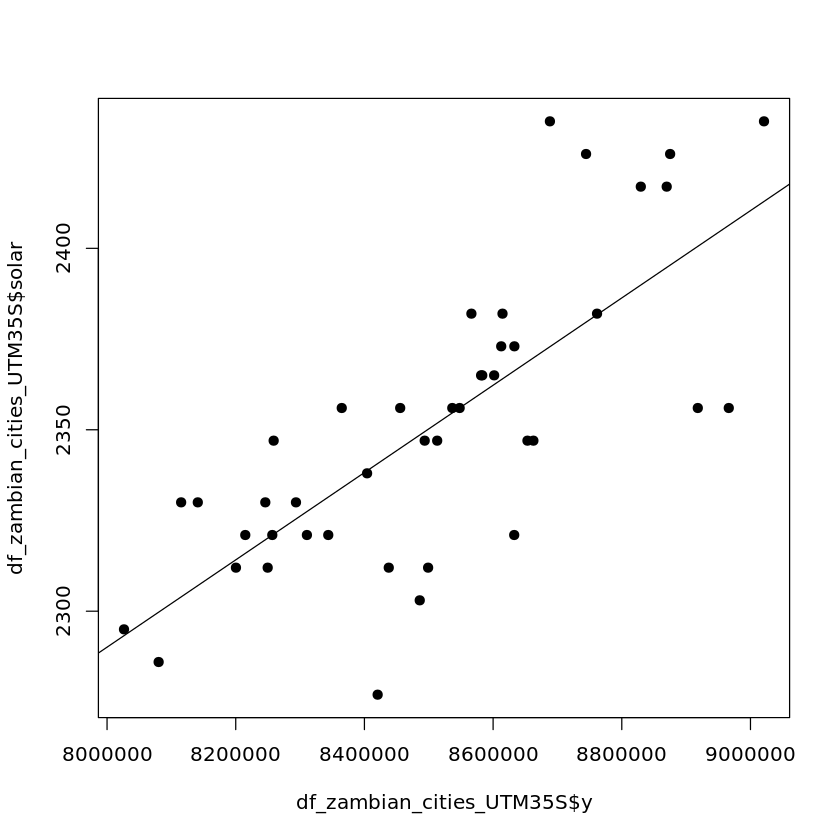
Latitude appears to have a stronger relationship with solar output which would be expected (why do you think this is?).
plot(df_zambian_cities_UTM35S$y, df_zambian_cities_UTM35S$solar, pch=19)
abline(lm(df_zambian_cities_UTM35S$solar ~ df_zambian_cities_UTM35S$y))
We’ll now create a model that predicts solar based on a 1st order linear regression against longitude/latitude.
trend_1st <- lm(solar~x+y, data=df_zambian_cities_UTM35S)
summary(trend_1st)
Call:
lm(formula = solar ~ x + y, data = df_zambian_cities_UTM35S)
Residuals:
Min 1Q Median 3Q Max
-63.083 -9.265 1.744 19.652 62.488
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.321e+03 1.605e+02 8.230 3.87e-10 ***
x -1.636e-06 1.804e-05 -0.091 0.928
y 1.212e-04 1.954e-05 6.203 2.45e-07 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 26.87 on 40 degrees of freedom
Multiple R-squared: 0.5649, Adjusted R-squared: 0.5432
F-statistic: 25.97 on 2 and 40 DF, p-value: 5.903e-08
We’ll now create a grid that we will carry out the predictions over
grid <- st_as_sf(rasterToPoints(solar, spatial=TRUE)) # uses existing raster cell centres as point locations
grid$solar <- NULL # clear existing data
plot(grid, pch=".")
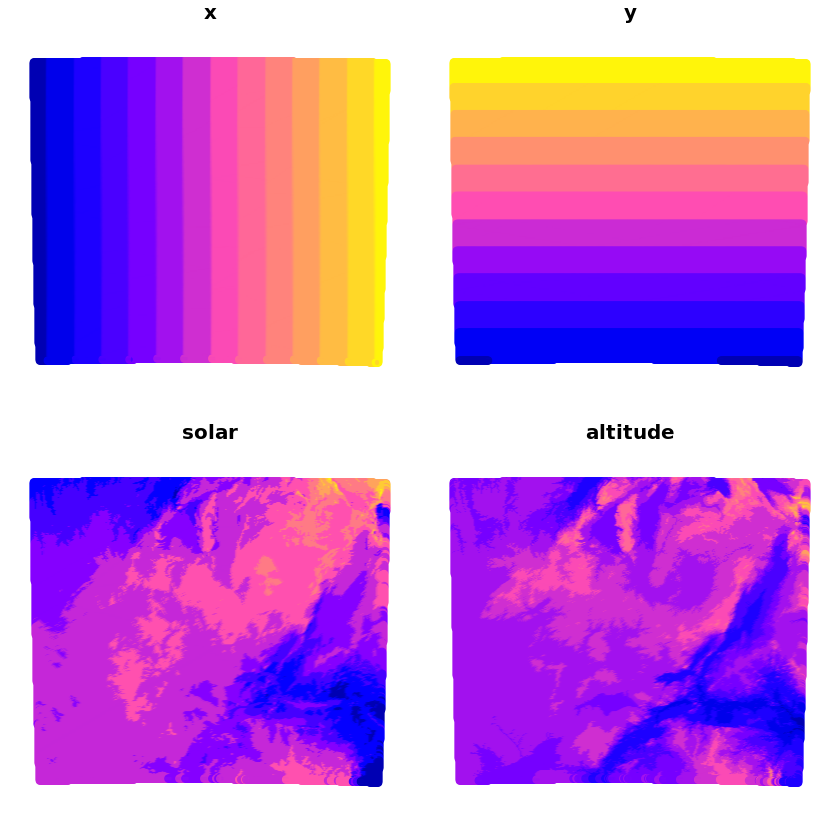
We’ll now populate this empty grid with the x/y values and solar/altitude values
grid_coords <- as.data.frame(st_coordinates(grid))
grid$x <- grid_coords[, 1]
grid$y <- grid_coords[, 2]
grid$solar <- extract(solar, grid_coords[, 1:2])
grid$altitude <- extract(altitude, grid_coords[, 1:2])
plot(grid)
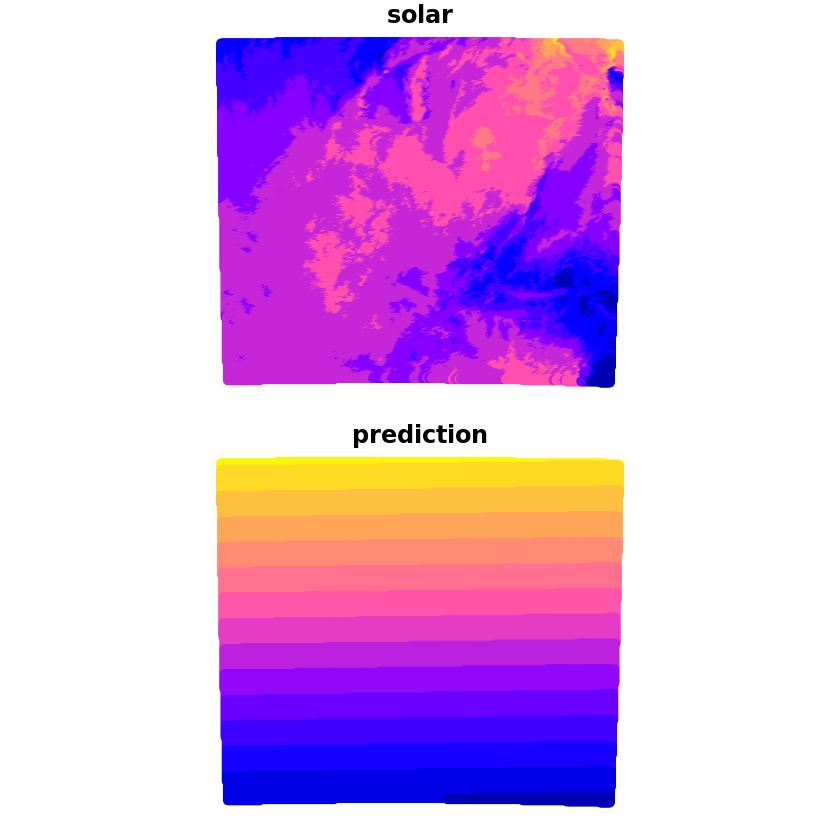
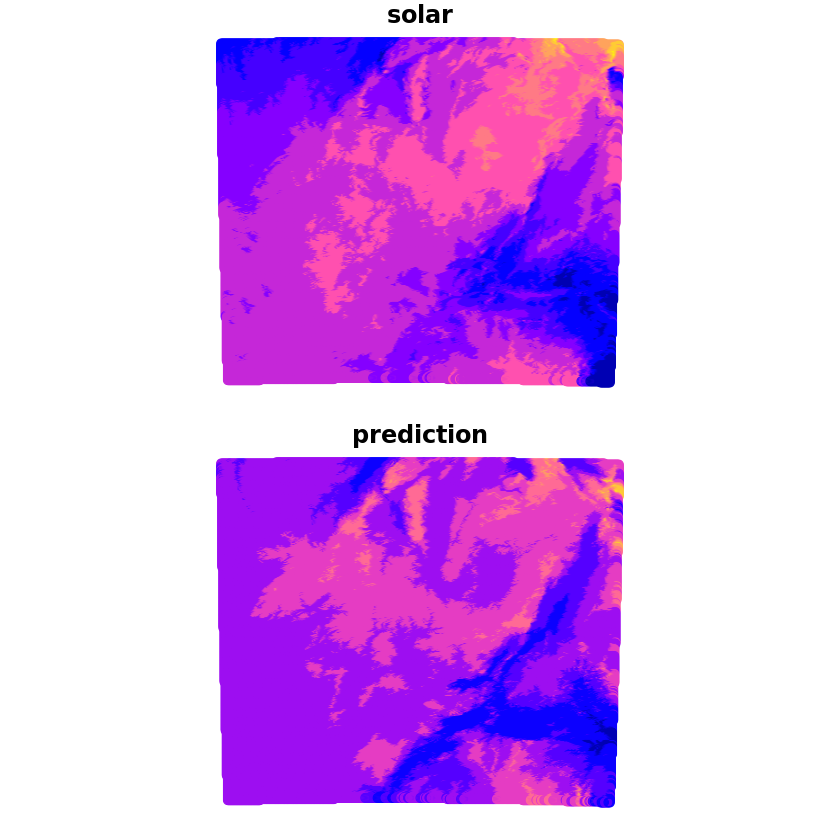
We can now make the prediction which we’ll compare with the true solar values, we can see that whilst it captures some of the wider characteristics there are a lot of discrepancies still.
grid_pred <- predict(trend_1st, newdata=grid, se.fit=TRUE)
grid$prediction <- grid_pred$fit
plot(grid[c('solar', 'prediction')])
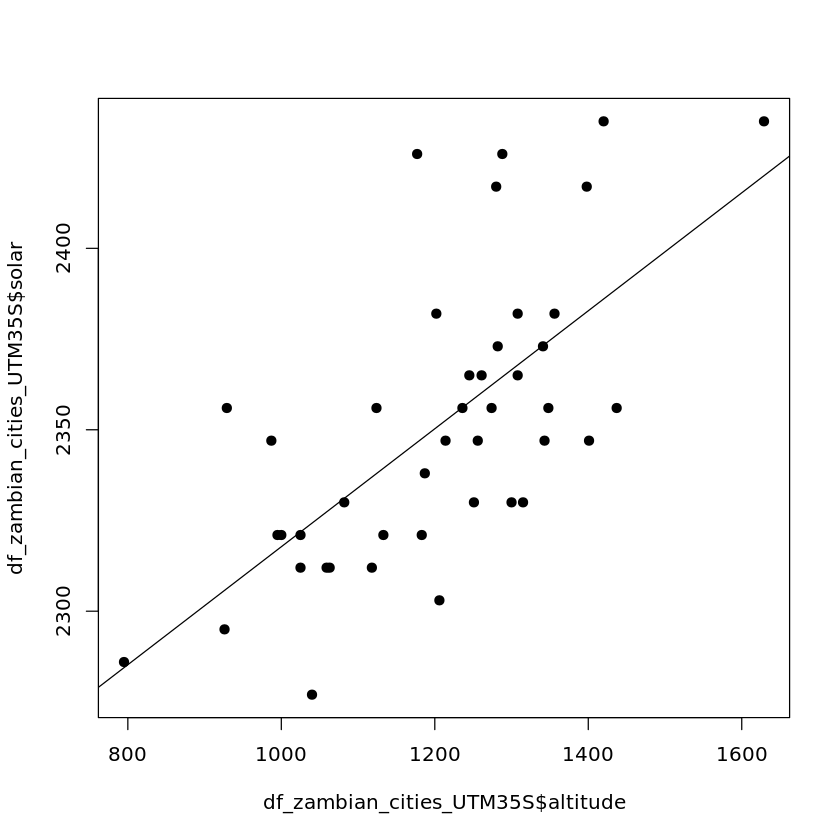
We don’t just have to regress against longitude/latitude though, here we’ll repeat the previous steps but only use latitude as a regressor.
First we’ll visualise the relationship between the two.
plot(df_zambian_cities_UTM35S$altitude, df_zambian_cities_UTM35S$solar, pch=19)
abline(lm(df_zambian_cities_UTM35S$solar ~ df_zambian_cities_UTM35S$altitude))
Then we’ll fit a model and use it in our prediction.
trend_1st <- lm(solar~altitude, data=df_zambian_cities_UTM35S)
grid_pred <- predict(trend_1st, newdata=grid, se.fit=TRUE)
grid$prediction <- grid_pred$fit
plot(grid[c('solar', 'prediction')])
Questions¶
Best Fit Model¶
What combination of the inputs produces the most accurate results?
Should we use the standard r2 score or the adjusted r2 score?
How do the results change when you use a test/train split?
How could we improve over a random sample on the test/train split? (Think about what you would do with time-series cross-validation)
#